Run NMF (non-negative matrix factorization)
Usage
RunNMF(object, ...)
# S3 method for class 'Seurat'
RunNMF(
object,
assay = NULL,
layer = "data",
features = NULL,
nbes = 50,
nmf.method = "RcppML",
tol = 1e-05,
maxit = 100,
rev.nmf = FALSE,
ndims.print = 1:5,
nfeatures.print = 30,
reduction.name = "nmf",
reduction.key = "BE_",
verbose = TRUE,
seed.use = 11,
...
)
# S3 method for class 'Assay'
RunNMF(
object,
assay = NULL,
layer = "data",
features = NULL,
nbes = 50,
nmf.method = "RcppML",
tol = 1e-05,
maxit = 100,
rev.nmf = FALSE,
ndims.print = 1:5,
nfeatures.print = 30,
reduction.key = "BE_",
verbose = TRUE,
seed.use = 11,
...
)
# Default S3 method
RunNMF(
object,
assay = NULL,
layer = "data",
nbes = 50,
nmf.method = "RcppML",
tol = 1e-05,
maxit = 100,
rev.nmf = FALSE,
ndims.print = 1:5,
nfeatures.print = 30,
reduction.key = "BE_",
verbose = TRUE,
seed.use = 11,
...
)Arguments
- object
An object. This can be a Seurat object, an Assay object, or a matrix-like object.
- ...
Additional arguments passed to RcppML::nmf or NMF::nmf function.
- assay
A character string specifying the assay to be used for the analysis. Default is NULL.
- layer
A character string specifying the layer name to be used for the analysis. Default is "data".
- features
A character vector specifying the features to be used for the analysis. Default is NULL, which uses all variable features.
- nbes
An integer specifying the number of basis vectors (components) to be computed. Default is 50.
- nmf.method
A character string specifying the NMF algorithm to be used. Currently supported values are "RcppML" and "NMF". Default is "RcppML".
- tol
A numeric value specifying the tolerance for convergence (only applicable when nmf.method is "RcppML"). Default is 1e-5.
- maxit
An integer specifying the maximum number of iterations for convergence (only applicable when nmf.method is "RcppML"). Default is 100.
- rev.nmf
A logical value indicating whether to perform reverse NMF (i.e., transpose the input matrix) before running the analysis. Default is FALSE.
- ndims.print
An integer vector specifying the dimensions (number of basis vectors) to print in the output. Default is 1:5.
- nfeatures.print
An integer specifying the number of features to print in the output. Default is 30.
- reduction.name
A character string specifying the name of the reduction to be stored in the Seurat object. Default is "nmf".
- reduction.key
A character string specifying the prefix for the column names of the basis vectors. Default is "BE_".
- verbose
A logical value indicating whether to print verbose output. Default is TRUE.
- seed.use
An integer specifying the random seed to be used. Default is 11.
Examples
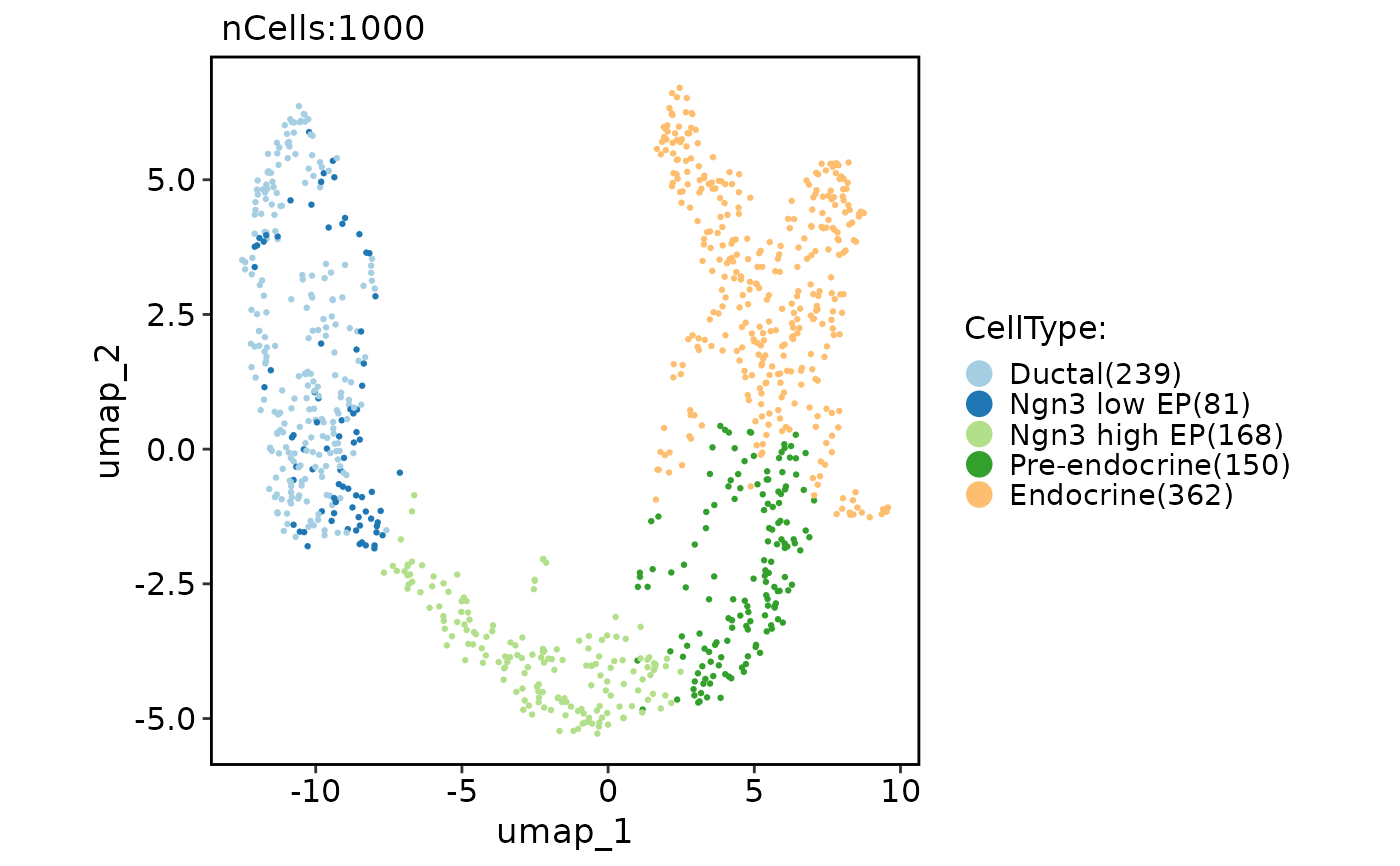
pancreas_sub <- RunNMF(object = pancreas_sub)
#> Error in check_R("zdebruine/RcppML"): could not find function "check_R"
CellDimPlot(pancreas_sub, group.by = "CellType", reduction = "nmf")
#> Warning: No shared levels found between `names(values)` of the manual scale and the data's fill values.